***Application Closed***
Shaping Authentic Practices by Engaging in Modeling of A Topic with Teachers to Explore Research in Science (SHAPE MATTERS) is funded by the NIH Science Education Partnership Award. The program has three objectives.
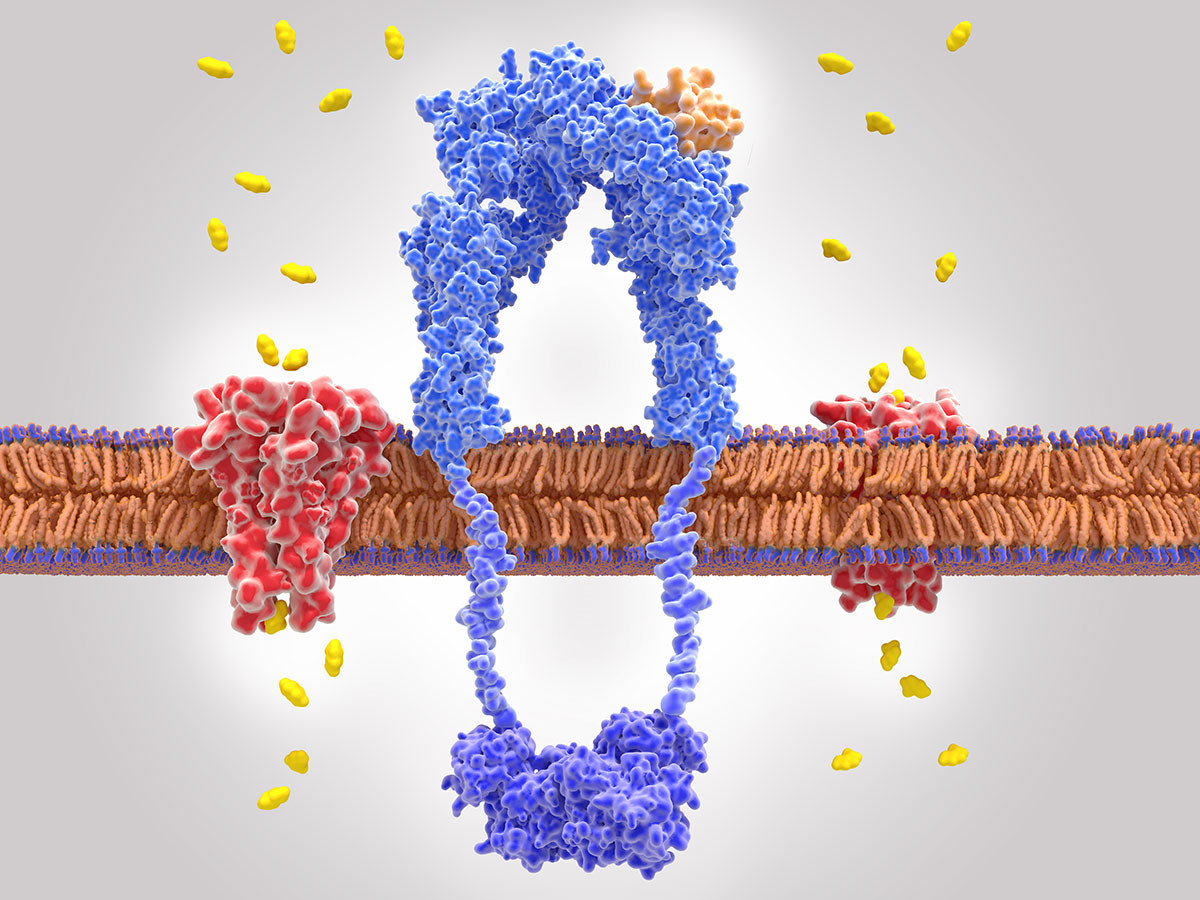
Description: Shaping Authentics Practices by Engaging in Modeling of A Topic with Teachers to Explore Research in Science (SHAPE MATTERS) is funded by the NIH Science Education Partnership Award. The program has three objectives.

Director Center for Science and the Schools, Associate Professor of Science Education
As the Director of the Center for Science and the Schools, Dr. Hill serves as the education lead on many technical grants across the STEM colleges at Penn State. In her role, she oversees the development, organization, management, and evaluation of education programs that serve to bridge cutting edge research to K-12 education. Her research focuses on three areas: a) designing professional development programs that engage teachers in the practices of scientists and engineers and promote effective strategies for engaging K-12 students in classroom research projects; b) examining teachers’ pedagogical content knowledge for supporting student-led research projects in the classroom; and c) building outreach programs that bridge the research of STEM faculty and graduate students with K-12 education. Prior to coming to Penn State, Kathy worked as an environmental scientist, science teacher, and Assistant Professor of Education at Bethany College in West Virginia.

STEM Education Outreach Specialist, Doctoral Student Curriculum and Instruction (Science Education)
Email: ams5306@psu.edu
Amber is a STEM Education Outreach Specialist at the Center for Science and the Schools, former high school biology teacher, and current doctoral student in Science Education at Penn State. In her current role at CSATS, she works with STEM research faculty to design and implement content-specific professional development for teachers that focuses on the practices of scientists and engineers at Penn State. As a doctoral student, her research focuses on a) the intersections of scientific modeling and computing in high school biology classrooms; and b) designing professional development programs that engage teachers in the practices of scientists and engineers. Before coming to Penn State, she taught Biology, Honors Biology, Physical Science, Anatomy and Physiology, and Marine Science for five years. While teaching, she also got involved in leading professional development for science teachers through collaborating with the Milwaukee School of Engineering Center for Biomolecular Modeling in her district and at local and national conferences.

Tiffany is a former high school science teacher at Middletown Area High School outside of Harrisburg, PA. In her seven years of teaching, she taught Earth Systems Science, Biology, Advanced Biology, AP Biology, and Forensics. She led building-level professional development sessions and presented at various educational conferences on incorporating research projects in the classroom. Tiffany’s main interest lies in introducing students to authentic science by helping teachers understand how to engage students in the practices of scientists and engineers. Her role at CSATS is to work with research faculty at Penn State to bring current research to the classroom by developing content-specific professional development for teachers. She also engages in building relationships with school districts in the greater Harrisburg area to better support science education.

Associate Professor of Biochemistry and Molecular Biology in the College of Medicine.
Dr. Ropson’s research has focused on the structure, folding, and function of proteins. He has examined the folding mechanism of ß-sheet proteins in the intracellular fatty acid binding protein family using kinetic studies of the fluorescence and circular dichroism spectra as the proteins move from the unfolded to the native state. These proteins have low levels of sequence homology, and there appears to be considerable differences in the path of folding for these sequences to the same final structure. Dr. Ropson has also worked on the effects of mutations on retroviral capsid assembly, and the adaptation of protein function to environmental stresses. He has been involved with teacher molecular structure and modeling to undergraduate, graduate, medical, and high school students.
Associate Professor of Chemistry and of Biochemistry and Molecular Biology at Penn State University Park.
Dr. Boal’s research focuses on understanding the structural differences between members of large metalloenzyme superfamilies that share common features buth promote different reactions or use distinct cofactors. Targets are unified in their ability to activity strong C-H, N-H, or O-H bonds. She characterizes stable reactant and product complexes with an increasing focus on development and implementation of crystallographic approaches to study metalloenzyme reaction intermediates.

Research Professor of Biochemistry and Molecular Biology, Director of X-Ray Crystallography Facility
Dr. Yennawar has collaborated with the research faculty from various departments at the Penn State University Park campus as well as Penn State’s sister campuses. Primarily, Dr. Yennawar has been involved in crystallization, single crystal X-ray data collection at home and synchrotron laboratories, determination and analyzing of three dimensional structures of molecules of interest using X-ray diffraction techniques (small and macromolecular). He has taught the Biomolecular Structure (BMMB 531) course for over a decade.
Associate Professor of Biochemistry and Molecular Biology, Director of the X-Ray Crystallography and Automated Biological Calorimetry Facilities at the Huck Institutes of the Life Sciences
Dr. Yennawar’s research focuses on three key areas at the cusp of materials and life sciences: (1) Atomic resolution structure determination by X-ray crystallography and micro-electron diffraction on a transmission cryo-electron microsome; (2) Designing protein, peptide, and DNA crystals for enhanced nonlinear optical properties for devices; and (3) Biophysical characterization by circular dichroism and transmission electron microscopy of the interfaced formed between DNA and synthetic materials for next generation solar cells and biosensors. She has held frequent training sessions and workshops on all of the biophysical equipment in the lab.

Professor of Chemistry, Professor of Biochemistry and Molecular Biology, Associate Department Head for Graduate Education
Biophysical chemistry is a discipline that merges an interest in the systems and processes from biochemistry with the principles and quantitative laws from physical chemistry. In the Showalter Laboratory, we make broad use of solution nuclear magnetic resonance (NMR) spectroscopy, in combination with thermodynamic analysis, chemical biology, and cellular assays to advance understanding of protein function. We place special emphasis on biophysical studies of macromolecular interactions involving partially disordered proteins. While we emphasize fundamental research, we are always mindful of our potential to advance molecular understanding of transcription and gene regulation, which we envision having sustained societal and biomedical impact.

Assistant Professor of Biochemistry and Molecular Biology
Proteins are regulated in many ways, including by binding of small molecules, other proteins and biological molecules. In many cases, these binding events result in structural changes across the protein that direct subsequent biological functions. Because these changes are often very subtle and can be invisible in crystal structures, it is not trivial to determine how protein motions are correlated with biological activity.
Our laboratory uses a computational method known as molecular dynamics simulations to investigate conformational dynamics in protein complexes and determine how these are altered in various functional states. We then use complementary biochemical and structural techniques to determine molecular mechanisms of long-range communication and signaling in proteins. By revealing how different conformational states are correlated with biological outcomes, we will develop strategies to control protein function for desirable applications. More specifically, we aim to understand the transcription factor FXR (farnesoid X receptor) and how its transcriptional activity is modulated by diverse small molecules. FXR controls critical metabolic genes, making it a therapeutic target for diseases such as diabetes and fatty liver disease. Members of this SMART team will explore the structure of FXR and predict how small molecules alter this structure to achieve distinct transcriptional outcomes.

Associate Professor of Biochemistry and Molecular Biology and Director of Core Facilities
Maria is a structural biologist who uses a range of biophysical tools to understand how proteins are regulated during their biological function. She has studied a range of molecular machines and circuits focusing on how they are precisely controlled during complex biological functions and how genetic mutations and species dependent sequence differences affect their function and substrate specificity on a local and organismal level. This project will be focused around control of carbohydrate metabolism in bacteria present in the human gut and is in collaboration with the Townsend and Flanagan labs in the College of Medicine in Hershey. In E. coli the CRP/CAP-cAMP control of lactose utilization is one of the most studied and important biological circuits with implications for both bacterial growth and for understanding the complex genetic mechanisms regulating the human gene expression. Specifically, we will focus on the “Master” regulator for carbohydrate metabolism in Bacteriodes thetaiotaomicron, a commensal human bacteria that contributes to weight gain in humans. This protein is a member of the CAP family, but it is not regulated by cAMP and its regulator is currently unknown. The SMART team will join us using structural, computational and biophysical tools to identify the regulator for this protein.

Associate Professor of Chemistry
Year: 2022-2023
The COVID-19 pandemic has highlighted the need to develop effective antiviral therapies for SARS-CoV-2 and related positive-strand RNA viruses. In the Boehr lab, we study the protein structure and dynamics of two common antiviral targets – the RNA-dependent RNA polymerase (RdRp) and the viral protease that cleaves the viral polyprotein into its component parts. Our work has highlighted the importance of understanding not just the static structure, but also how motions within the protein impact function and regulation. We propose that just as everyday machines have moving parts to function, so do these “protein nanomachines.” More specifically, our work has shown that amino acid substitutions distant from the active site of the RdRp change error frequency not by changing the static protein structure but by modulating the internal motions within the RdRp. These functional changes have implications for viral pathogenesis and vaccine development. Antiviral nucleotides can also impede these functionally-important motions to stop RdRp function. Teachers will explore the structure of this well-conserved and important antiviral target, but also gain an appreciation for the wide variety of motions and structural changes that ultimately drive protein function.

Professor of Biochemistry and Molecular Biology
Year: 2021-2022
The DNA of eukaryotic chromatin and chromosomes is repeatedly coiled around histone octamers forming “beads-on-a-string” chains of nucleosomes connected by extended DNA linkers. The linker DNA is additionally folded by linker histone H1 and other architectural chromatin proteins. Understanding how do nucleosome chains fold and unfold to regulate DNA transcription, replication, repair, and overall compaction in living cells is currently a major research goal in our laboratory. Our work on a developmentally regulated chromatin-condensing factor, MENT, uncovered the first chromatin architectural protein for which the mechanism of its action emerged from biochemical, electron microscopic, and X-ray crystal structure analyses. Our more recent studies using the unique electron microscopy-based technique, EMANIC (EM-assisted nucleosome interaction capture) helped to change the previously established paradigm of the regular chromatin fiber coiling and suggested a mechanism of the heteromorphic nucleosome zigzag folding by histone H1. Furthermore, our advance in understanding the basic principles of chromatin structure has a direct application to biomedical problems. For example, our collaboration with Penn State University Park scientists demonstrated that global nucleosome chain unfolding underlies NETosis, a process which mediates bacterial trapping by neutrophils of human blood but also leads to acute inflammation, thrombosis and other life-threatening disorders. We are now studying the mechanism of global chromatin unfolding using a combination of cryo-electron microscopic tomography with 3-dimensional modeling based on nucleosome structures available from the Protein Data Bank. Members of this SMART team will generate different models of nucleosomes with and without linker histone H1. This work will help us to test a hypothesis that an inhibition of histone H1-mediated linker DNA folding is instrumental in driving the general unfolding of the nucleosome chain leading to NETosis.

MS, PhD Professor, Department of Cellular and Molecular Physiology
Year: 2021-2022
The laboratory of Dr. Christopher Yengo studies molecular motor and cytoskeletal proteins and their role in human disease. The lab is currently focused on myosin motors and the actin cytoskeleton. Biochemical, biophysical, and cell biological approaches are utilized to investigate basic molecular mechanisms of motor-based transport, contraction and organization of the actin cytoskeleton. In the long term, the Yengo lab hopes to develop therapeutic strategies for diseases that impact actomyosin-based functions.

Year: 2021-2022
The Murakami Lab is in need of motivated graduate students and postdoctoral researchers that are interested in eukaryotic transcription and structural studies.

Year: 2021-2022
Research in the Bevilacqua Lab is centered on understanding functions of RNA in nature at the molecular level. RNA is a fascinating molecule because it has both genetic and functional capabilities. This has led to the notion that RNA was particularly important in the emergence of life on Earth–the “RNA World Hypothesis”. RNA is also of interest because it is involved in a wide range of important biological pathways and can be used as both a biological target and a chemical tool. We work on RNAs ranging from simple model systems to the entire transcriptome (tens of thousands of RNAs) in living organisms. There are three main projects in the lab broadly defined: ribozyme mechanism, RNA folding in vivo, and roles RNA may have played in the emergence of life on early earth. Experimental approaches range from RNA molecular biology and genomics to biophysics, and we often develop new methods in these areas to facilitate the research. We collaborate with outstanding scientists including biologists, physicists, and theorists.

Assistant Professor of Biochemistry and Molecular Biology
Year: 2021-2022
The Swulius lab uses cryo-electron tomography (cryo-ET) to capture snapshots of living cells at molecular resolution. Our goal is to understand how nanoscale changes in the underlying cytoskeleton are translated into dynamic cellular-level behavior. Currently we are focused on actin filaments and the proteins that regulate their assembly and structure in the tips of developing neuronal processes. We are looking at the interplay between two different actin binding proteins known as fascin and cofilin, and how they function to alter the structure and physical properties of actin networks during neuronal outgrowth. Teachers will work with cryo-ET data to explore and model the structure of actin filament networks for quantitative analysis.
The SHAPE MATTERS program will offer a 10-day professional development workshop for teachers during the summer occurring on the Penn State University Park campus. The professional development is geared towards secondary life science and chemistry teachers. During the professional development, teachers will collaborate with science education faculty from the Center for Science and the Schools and research scientists from the College of Medicine and the Eberly College of Science. To explore molecular stories related to human health, teachers will engage in research techniques such as crystallization, structure determination, and modeling using the molecular visualization software, Jmol. Teachers will work with the SHAPE MATTERS team to co-construct a molecular modeling research project for their classroom. The research project will support student participation in the SMART teams program.
Secondary life science and chemistry teachers
The application requires general contact information, direct supervisor information, an up-to-date resume, 4 short essay questions, 3 professional references, and a support letter from your immediate supervisor.
Nearly half of the antibiotics that are currently prescribed target the bacterial ribosome. However, bacteria are rapidly evolving new ways to resist these antibiotics. One emerging mode of enzymatic antibiotic resistance is the structural modification of the ribosome by the radical SAM enzyme Cfr. Radical SAM (RS) enzymes share a common functional motif, an iron-sulfur cluster that coordinates S-adenosylmethionine (SAM), to generate a 5’-deoxyadenosine radical (5’-dA•). The 5’-dA• moiety in RS proteins initiates reactions with substrates by abstracting an H-atom. After Cfr performs this chemically challenging first step, several other steps occur, culminating in the methylation (i.e., the formation of a carbon-carbon bond) of ribosomal ribonucleic acid (RNA). Cfr uses this powerful chemistry to methylate a single site in the bacterial ribosome, the eighth carbon of the adenine ring on A2503. This structural modification inhibits antibiotic binding to the bacterial ribosome conferring antibiotic resistance. In collaboration with Squire Booker’s laboratory, our research focuses on structural determination of Cfr and a related radical SAM methylase, RlmN, to understand how Cfr and RlmN recognize substrate and perform the chemistry associated with RNA modification. To date, we have solved structures of an RNA-bound RlmN intermediate, the first view of a radical SAM enzyme in complex with a large macromolecular substrate.

In protein biochemistry, it is commonly assumed that a protein’s folded state determines its function. However, data emerging over the past decade conclusively show that approximately one-third of all human protein is not folded and yet these segments of “intrinsically disordered protein” often play essential roles in mediating protein-protein interactions. Thus, the premise motivating my laboratory’s research is that folding is not required to impart unique structural features in proteins that drive their biological functions. For example, the transcription factor Pdx1 regulates the production of insulin in pancreatic β-cells. Accomplishing this crucial function requires reversible formation of protein-protein complexes that either activate or repress Pdx1-stimulated transcription. In this project, the SMART team will explore how the intrinsically disordered C-terminal region of Pdx1 interacts with its negative regulator SPOP, which happens in order to avoid cellular stress associated with unnecessary insulin production when blood glucose levels are low. Analysis of co-crystal structures of Pdx1 peptides bound to SPOP will reveal how disordered proteins with diverse amino acid sequences can adopt functionally equivalent structures when bound to a folded partner. Perhaps unsurprisingly, the pdx1 gene is often mutated in diabetic patients; members of this SMART team will formulate a structure-based hypothesis for the mechanism whereby one diabetes-associated Pdx1 mutant may drive a disease phenotype by disrupting Pdx1-SPOP interactions.

Proteins are regulated in many ways, including by binding of small molecules, other proteins and biological molecules. In many cases, these binding events result in structural changes across the protein that direct subsequent biological functions. Because these changes are often very subtle and can be invisible in crystal structures, it is not trivial to determine how protein motions are correlated with biological activity.
Our laboratory uses a computational method known as molecular dynamics simulations to investigate conformational dynamics in protein complexes and determine how these are altered in various functional states. We then use complementary biochemical and structural techniques to determine molecular mechanisms of long-range communication and signaling in proteins. By revealing how different conformational states are correlated with biological outcomes, we will develop strategies to control protein function for desirable applications. More specifically, we aim to understand the transcription factor FXR (farnesoid X receptor) and how its transcriptional activity is modulated by diverse small molecules. FXR controls critical metabolic genes, making it a therapeutic target for diseases such as diabetes and fatty liver disease. Members of this SMART team will explore the structure of FXR and predict how small molecules alter this structure to achieve distinct transcriptional outcomes.

Maria is a structural biologist who uses a range of biophysical tools to understand how proteins are regulated during their biological function. She has studied a range of molecular machines and circuits focusing on how they are precisely controlled during complex biological functions and how genetic mutations and species dependent sequence differences affect their function and substrate specificity on a local and organismal level. This project will be focused around control of carbohydrate metabolism in bacteria present in the human gut and is in collaboration with the Townsend and Flanagan labs in the College of Medicine in Hershey. In E. coli the CRP/CAP-cAMP control of lactose utilization is one of the most studied and important biological circuits with implications for both bacterial growth and for understanding the complex genetic mechanisms regulating the human gene expression. Specifically, we will focus on the “Master” regulator for carbohydrate metabolism in Bacteriodes thetaiotaomicron, a commensal human bacteria that contributes to weight gain in humans. This protein is a member of the CAP family, but it is not regulated by cAMP and its regulator is currently unknown. The SMART team will join us using structural, computational and biophysical tools to identify the regulator for this protein.

Year: 2021-2022
Research in the Bevilacqua Lab is centered on understanding functions of RNA in nature at the molecular level. RNA is a fascinating molecule because it has both genetic and functional capabilities. This has led to the notion that RNA was particularly important in the emergence of life on Earth–the “RNA World Hypothesis”. RNA is also of interest because it is involved in a wide range of important biological pathways, recognized most recently as the genetic material of the SARS-CoV-2 virus and the basis of the Moderna and Pfizer mRNA vaccines. We work on RNAs ranging from simple model systems to the entire transcriptome (tens of thousands of RNAs) in living organisms. The project our teachers will be working on will involve recognition of ligands by riboswitches. Riboswitches are a recently discovered class of RNA molecules that specifically and tightly bind small molecules to regulate gene expression. Our teachers will study two closely related purine riboswitches and understand the molecular basis for their specificity. Teachers will learn chemical concepts interfacing with physics and biology, including folding of RNA, molecular recognition of ligands, and regulation of gene expression.

Year: 2021-2022
The ongoing COVID-19 pandemic that is threatening people's lives across the world is underscoring the urgent need to develop effective antiviral agents for SARS-CoV-2, the causative agent of COVID-19. The SARS-CoV-2 main protease Mpro (its alternate name 3C-like protease 3CLpro), which cleaves the viral non-structural polyprotein (NSP) for assembling a replication-transcription complex (RTC), is one of excellent antiviral targets to prevent SARS-CoV-2 replication. The Murakami’s group has been determining the atomic-resolution X-ray crystallographic structure of Mpro in complex with protease inhibitor (Figure below) for understanding the mechanisms of Mpro inhibition by small molecules and for developing new SARS-CoV-2 agents.

Year: 2021-2022
The laboratory of Dr. Christopher Yengo studies molecular motor and cytoskeletal proteins and their role in human disease. The lab is currently focused on myosin motors and the actin cytoskeleton. Biochemical, biophysical, and cell biological approaches are utilized to investigate basic molecular mechanisms of motor based transport, contraction, and organization of the actin cytoskeleton. In the long term, the Yengo lab hopes to develop therapeutic strategies for diseases that impact actomyosin based functions. Specific diseases being investigated are inherited forms of heart disease and deafness.

Year: 2021-2022
The COVID-19 pandemic has highlighted the need to develop effective antiviral therapies for SARS-CoV-2 and related positive-strand RNA viruses. In the Boehr lab, we study the protein structure and dynamics of two common antiviral targets – the RNA-dependent RNA polymerase (RdRp) and the viral protease that cleaves the viral polyprotein into its component parts. Our work has highlighted the importance of understanding not just the static structure, but also how motions within the protein impact function and regulation. We propose that just as everyday machines have moving parts to function, so do these “protein nanomachines.” More specifically, our work has shown that amino acid substitutions distant from the active site of the RdRp change error frequency not by changing the static protein structure but by modulating the internal motions within the RdRp. These functional changes have implications for viral pathogenesis and vaccine development. Antiviral nucleotides can also impede these functionally-important motions to stop RdRp function. Teachers will explore the structure of this well-conserved and important antiviral target, but also gain an appreciation for the wide variety of motions and structural changes that ultimately drive protein function.

Year: 2021-2022
The DNA of eukaryotic chromatin and chromosomes is repeatedly coiled around histone octamers forming “beads-on-a-string” chains of nucleosomes connected by extended DNA linkers. The linker DNA is additionally folded by linker histone H1 and other architectural chromatin proteins. Understanding how do nucleosome chains fold and unfold to regulate DNA transcription, replication, repair, and overall compaction in living cells is currently a major research goal in our laboratory. Our work on a developmentally regulated chromatin-condensing factor, MENT, uncovered the first chromatin architectural protein for which the mechanism of its action emerged from biochemical, electron microscopic, and X-ray crystal structure analyses. Our more recent studies using the unique electron microscopy-based technique, EMANIC (EM-assisted nucleosome interaction capture) helped to change the previously established paradigm of the regular chromatin fiber coiling and suggested a mechanism of the heteromorphic nucleosome zigzag folding by histone H1. Furthermore, our advance in understanding the basic principles of chromatin structure has a direct application to biomedical problems. For example, our collaboration with Penn State University Park scientists demonstrated that global nucleosome chain unfolding underlies NETosis, a process which mediates bacterial trapping by neutrophils of human blood but also leads to acute inflammation, thrombosis and other life-threatening disorders. We are now studying the mechanism of global chromatin unfolding using a combination of cryo-electron microscopic tomography with 3-dimensional modeling based on nucleosome structures available from the Protein Data Bank. Members of this SMART team will generate different models of nucleosomes with and without linker histone H1. This work will help us to test a hypothesis that an inhibition of histone H1-mediated linker DNA folding is instrumental in driving the general unfolding of the nucleosome chain leading to NETosis.

Year: 2021-2022
The Swulius lab uses cryo-electron tomography (cryo-ET) to capture snapshots of living cells at molecular resolution. Our goal is to understand how nanoscale changes in the underlying cytoskeleton are translated into dynamic cellular-level behavior. Currently we are focused on actin filaments and the proteins that regulate their assembly and structure in the tips of developing neuronal processes. We are looking at the interplay between two different actin binding proteins known as fascin and cofilin, and how they function to alter the structure and physical properties of actin networks during neuronal outgrowth. Teachers will work with cryo-ET data to explore and model the structure of actin filament networks for quantitative analysis.
